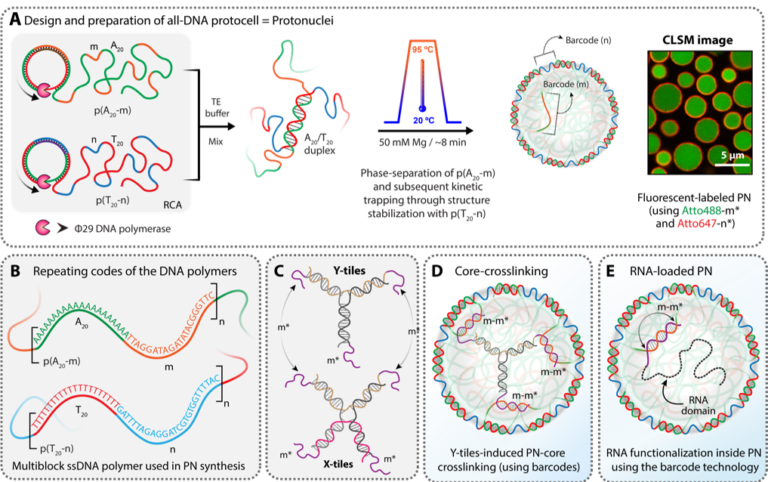
The long-term goal of this project is to establish biopolymer-based protonuclei compartments as a viable nucleus-mimicking platform for studying the assembly and molecular function of biological entities, while including effects such as molecular crowding and tuneable viscoelastic properties, together with fast analytical capabilities from the polymer colloid field. In the first phase of this project, we will develop and establish DNA- and RNA-containing “protonuclei” (PN) with tuneable soft hydrodynamic confinement properties as a system that will enable us to study and better understand phase separation of arginine and glycine (RG)-rich proteins, prototype phase-separating proteins with DNA/RNA-binding capacity. This class of protein is particularly important in DNA/RNA processing, for example, in transcription, DNA damage signalling, pre-mRNA splicing and mRNA translation. RG-rich regions mediate nucleic acid binding and protein–protein interactions, and often drive phase separation and assembly of membrane-less organelles (MLOs), for example, stress granules (SGs) or P granules. To gain new insights into the biological functions and regulation of the RG-rich proteome, we will use bioinformatics methods to classify RG-rich proteins in the proteome based on R/G ratio, the presence of other amino acids, post-translational modifications (PTMs), and the structures and short motifs present in RG-rich regions. We will analyse the conservation of RG-rich regions in homologues to point to functionally relevant regions. For human proteins, we will also collect data on disease mutations and protein interactions. Based on this analysis, we will select candidate RG-rich model proteins for further analysis. We will study the phase separation of RG-rich model proteins and peptides in the presence of single-stranded DNA (ssDNA) and RNA in vitro, in cellulo and “in protonucleo“. Within four years we aim to have established in protonucleo as an intermediate experimental platform between in vitro and in cellulo. Moreover, we expect to gain significant insights into the biological function and phase separation of the RG-rich proteome.
Related publications:
- R. Merindol, S. Loescher, A. Samanta, A. Walther*, Pathway-Controlled Formation of Mesostructured all-DNA Colloids and Superstructures. Nat. Nanotechnol. 13, 730 (2018).
- A. Samanta, V. Sabatino, T. R. Ward*, A. Walther*, Functional and Morphological Adaptation in DNA Protocells via Signal Processing Prompted by Artificial Metalloenzymes. Nat. Nanotechnol. 15, 914 (2020).
- P. Mier*, L. Paladin, S. Tamana, S. Petrosian, B. Hajdu-Soltész, A. Urbanek, A. Gruca, D. Plewczynski, M. Grynberg, P. Bernadó, Z. Gáspári, C. A. Ouzounis, V. J. Promponas, A. V. Kajava, J. M. Hancock, S. C. E. Tosatto, Z. Dosztanyi, M. A. Andrade-Navarro, Disentangling the complexity of low complexity proteins. Brief. Bioinform. 21, 458 (2020).
- M. Kamel, P. Mier, A. Tari, M. A. Andrade-Navarro*, Repeatability in protein sequences. J. Struct. Biol. 208, 86 (2019).
- S. Hutten*, S. Usluer, B. Bourgeois, F. Simonetti, H. M. Odeh, C. M. Fare, M. Czuppa, M. Hruska-Plochan, M. Hofweber, M. Polymenidou, J. Shorter, D. Edbauer, T. Madl, D. Dormann*, Nuclear Import Receptors Directly Bind to Arginine-Rich Dipeptide Repeat Proteins and Suppress Their Pathological Interactions. Cell Rep. 33, 108538 (2020).
- M. Hofweber#, S. Hutten#, B. Bourgeois, E. Spreitzer, A. Niedner-Boblenz, M. Schifferer, M.-D. Ruepp, M. Simons, D. Niessing, T. Madl, D. Dormann*, Phase Separation of FUS Is Suppressed by Its Nuclear Import Receptor and Arginine Methylation. Cell 173, 706 (2018).



