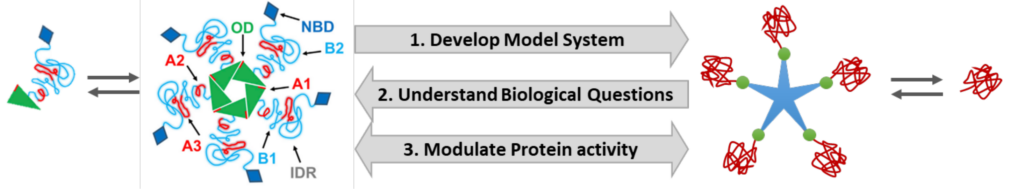
Proteins in nature exhibit diverse structures and sizes, some preferring to exist as monomers, while others form oligomers, ranging from dimers to larger clusters. Remarkably, the function of a protein is intrinsically linked to its structural state. Nature exploits this phenomenon to precisely regulate protein function, i.e. activation-deactivation or change in its function. Despite recognizing the effects of oligomerization on protein function, the intricate mechanisms involving multivalency, conformation, and flexibility, remain elusive. This knowledge gap is particularly pronounced for multifunctional proteins like some chaperones, transcription factors, or Nucleophosmin (NPM1). NPM1 predominantly adopts a pentameric state and is distributed between nucleolus and cytoplasm. NPM1 plays essential roles in mRNA transport, chromatin remodeling, apoptosis, and genome stability and shifts between monomer, pentamer, decamer as well as various supramolecular states, based on its specific function. To address this challenge, we propose a macromolecular chemistry approach aligned with SFB 1551’s mission, to achieve precise and switchable protein multivalency, oligomerization, and control of the associated phase transitions. This approach combines covalent and non-covalent designs, leveraging dendrimers and host-guest interactions. Our research will commence with a dendrimer-based model system to systematically investigate the impact of oligomerization on the multivalency, conformation, and flexibility of proteins. Subsequently, we’ll explore changes in phase separation dynamics across different oligomeric states, offering insights into cellular processes and protein regulation. Further, to control protein function within our model system, we propose a photocaging approach for photo-triggered, precise, and temporally programmable protein release from oligomers. Ultimately, our goal is to employ a supramolecular host-guest approach with multi-stimuli-responsive switchable protein oligomerization, with potential applications in cellular systems. This study promises to contribute to a deeper understanding of protein-protein interactions and the fine-tuning of cellular functions.
Johannes Gutenberg University Mainz

