Block co-polymers are comprised of multiple homopolymer regions that enable the design of polymer materials with sophisticated properties. For instance, the ability to form micelles can be realized by designing a di-block co-polymer of a hydrophobic and a hydrophilic sequence, leading to a core–shell structure in which hydrophilic
blocks entropically stabilize small structures and shield the hydrophobic core. However, polymer nanoassemblies
are comprised of a limited number of molecules, which implies they are susceptible to fluctuations.
These fluctuations can drive behaviour that cannot be well approximated by the law of large numbers. In
contrast to microscopically visible large assemblies, studies on the function of nano-assemblies in biology and
the dynamics and conformational states of the molecules involved are scarce, in large part due to technical
challenges. In this project, we pair polymer theory with experimental methods to investigate the properties and
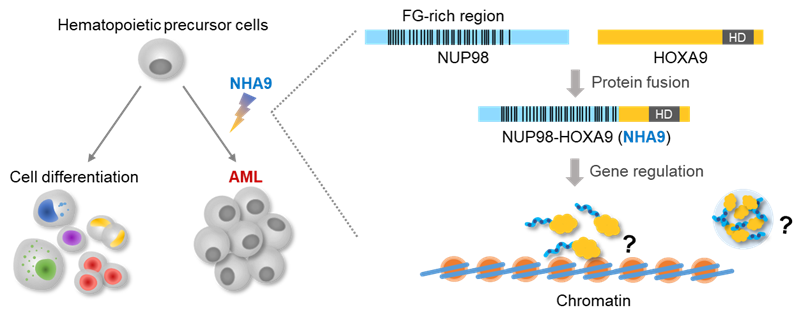
function of disordered protein domains fused with transcription factors, and how these can tune gene
regulation. We will investigate the extent to which such complex molecules can be described as sophisticated
multi-block co-polymers, and study how the individual blocks and their sum tune function. This will be
accompanied by a systematic bottom-up study to determine the design principles common to simple block copolymers
that enable the formation of stable nanoclusters of protein assemblies in solution. For the
transcription factor fusion proteins, we will also study their active state when surface bound to DNA. Our studies
will pair in vitro assays aimed at classifying and benchmarking solution and surface-mediated nano-assembly
processes with in vivo quantitative high-resolution fluorescence methods to determine the number, spatial
distribution and actual conformational states of the participating biopolymers inside living cells. Together this
will provide access to a particularly important regime in cellular signalling and transcriptional regulation by
investigating small “multi-block co-polymer clusters”, which are larger than tetramers but smaller than 1000-
mers.